LISI ULisboa Herbarium Sheet Mass Digitisation
Table of contents
Overview
This workflow illustrates part of the procedures adopted by a small university herbarium (about 80.000 specimens) which started the complete digitization and imaging of its herbarium sheets just a few years ago. The team in charge of this project is also reduced, consisting of a curator (part time), IT specialist (part time), digitizer/database operator and herbarium technician. The previous collaboration of an additional technical assistant, who started a prototype database in FileMaker Pro 3.x is also noteworthy. The collection database is presently managed using Specify 6. Regarding the hardware, the herbarium is equipped with an imaging station, consisting of an all-in-one PC, connected to a wireless Zebra barcode reader, one planetary scanner IS2 eScan, external drive (1Tb) and a Zebra carbon label printer. The resulting dataset, including (at the moment) over 76.000 records and more than 8.600 images, is available through GBIF.
The workflow is an adaptation to the local settings of a sequence of workflows published by Nelson et al. (2015), developed by the Dititization Group of iDigBio, available through GitHub and was prepared in the framework of the participation of Instituto Superior de Agronomia in the Research Infrastructure PORBIOTA.

Workflow
The presented diagrams is composed of six steps:
- Pre-Digitization Curation
- Digital Imaging Preparation
- Imaging Capture
- Imaging Processing
- Imaging Archiving
- Post-digitization Curation
The detailed description of the workflow is available here.
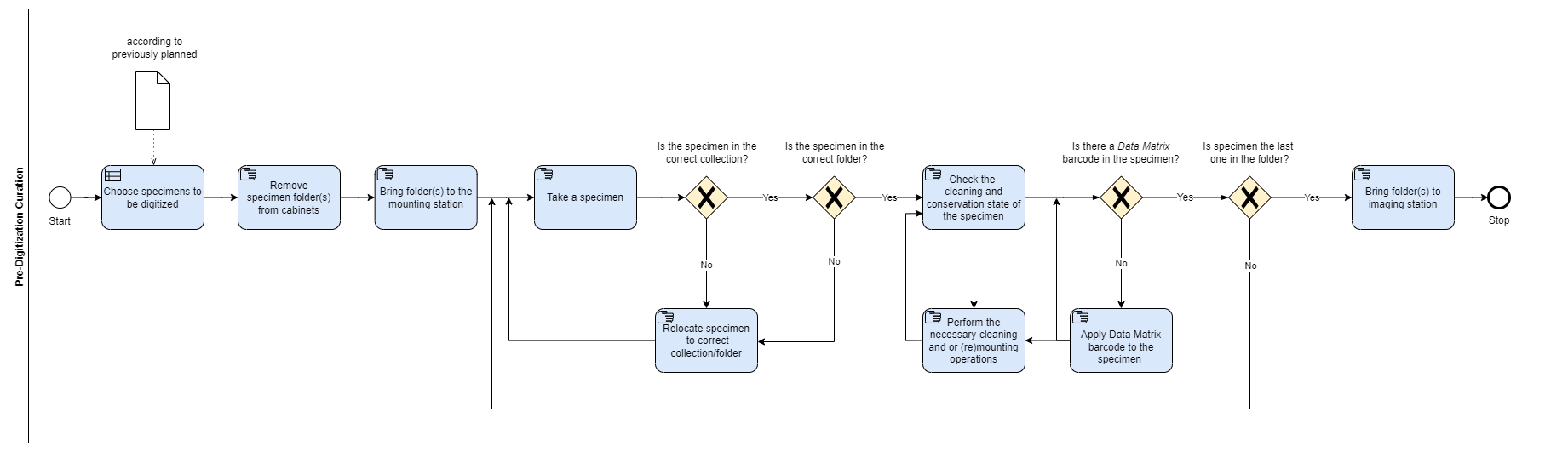
Pre-Digitization Curation
The ‘Pre-digitization Curation’ step involves tasks that occur prior to databasing or imaging, such as checking the cleaning and maintenance state of the material; placing the specimen Data Matrix barcodes; and transporting the specimens to the digitization station.
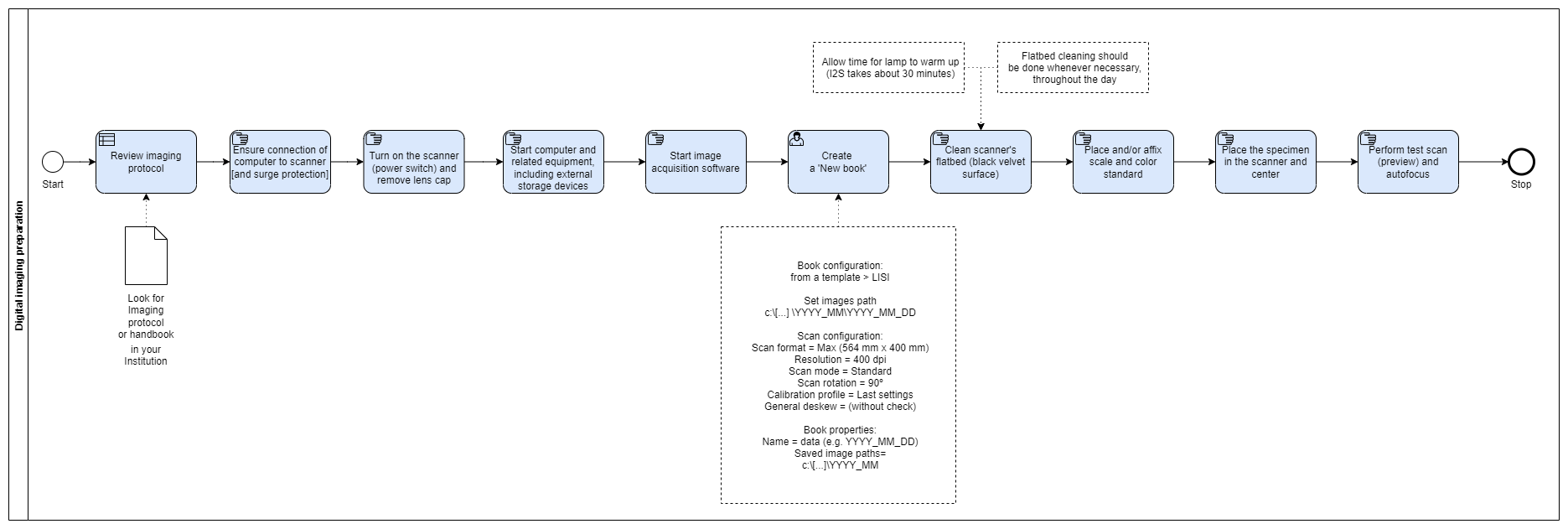
Digital Imaging Preparation
The Digital Imaging preparation is part of the ‘Digital Image Workflow’ and involves tasks destined to initiate and set up the imaging station, preparing it for sequential imaging of herbarium flat sheets.
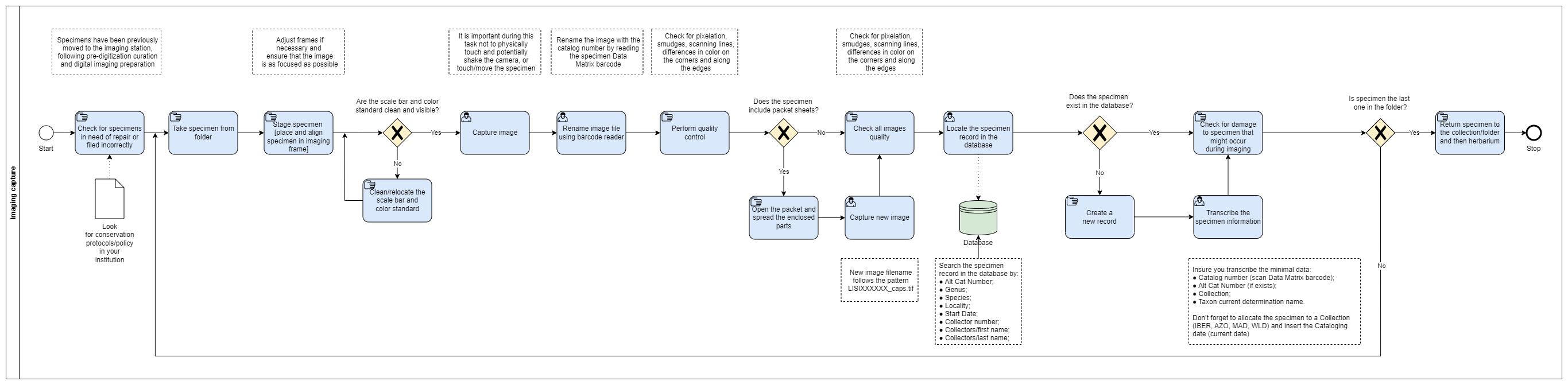
Imaging Capture
The ‘Image Capture workflow’ is subsequent to ‘Digital Imaging preparation’ workflow and is an ordered set of steps through which a specimen sheet is removed from an enclosing folder, imaged, returned to the original folder for refiling, and the resulting image examined for targeted quality. Specimens should also be checked for damage before and after imaging.
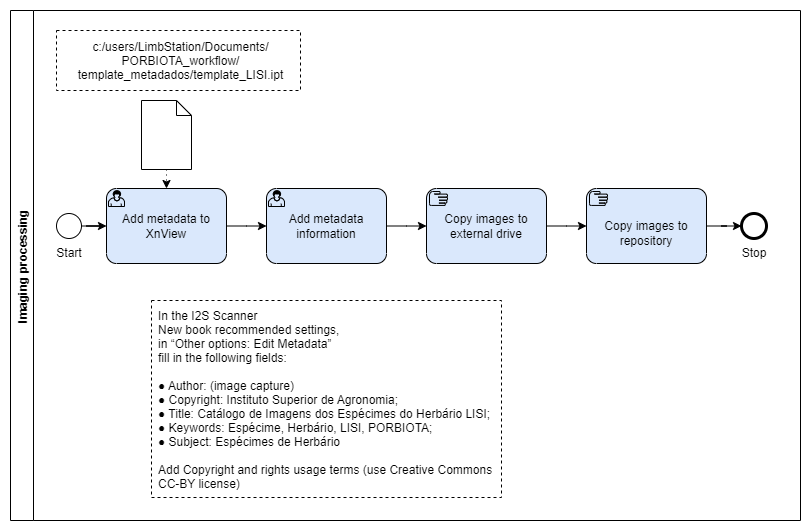
Imaging Processing
The Image Processing step involves completing image metadata with additional information not included at the capture and copying image files to a file server.
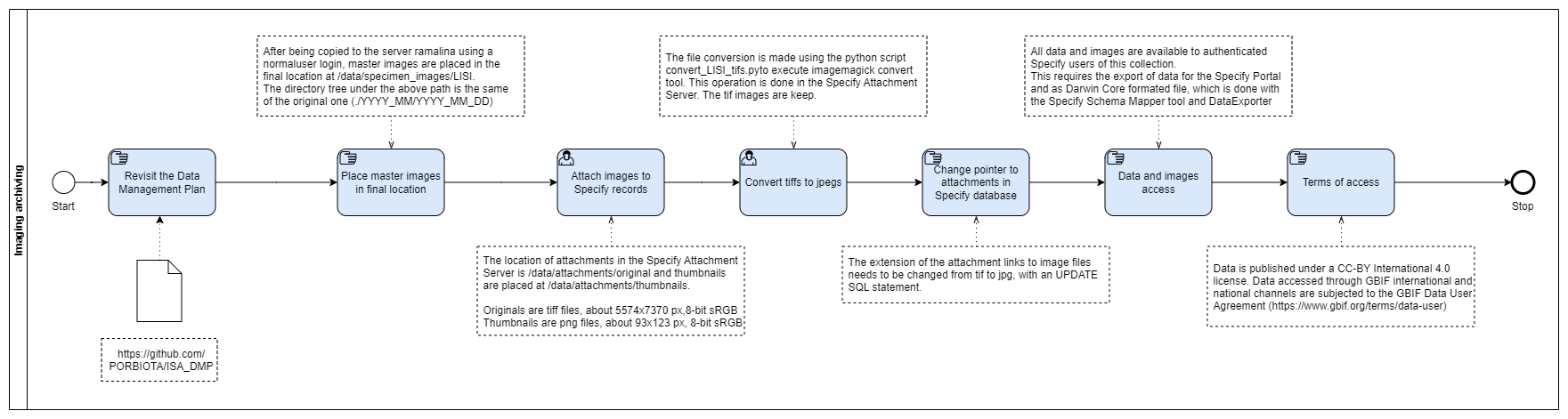
Imaging Archiving
The Imaging Archiving steps serve to place files in the final archiving location, and to include them to the collection management system (CMS, Specify 6) as an attach. The format of the attached files is jpeg, to ensure usability in local and web applications. The original tiff images are stored locally and backed up to a cloud infrastructure service.
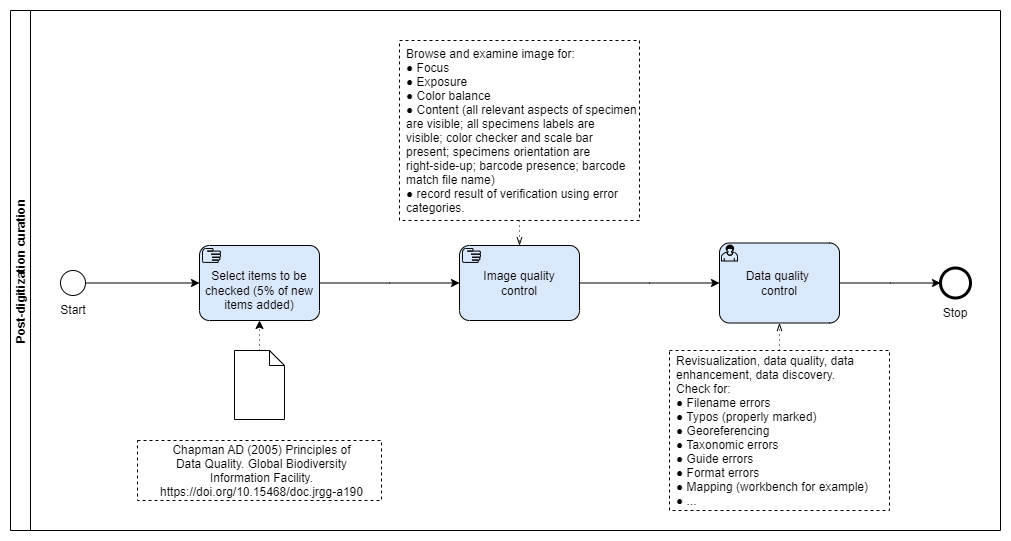
Post-digitization Curation
A quality control assessment is regularly performed at the CMS level, on a random sample of the new items added, to check for specimen data and images. Procedures follow recommendations by Champman (2005).
Resources and Requirements
This workflow is adapted to a small collections with the following available resources:
Personal
- a curator (part time)
- IT specialist (part time)
- digitizer/database operator
Equipment
Hardware
- imaging station:
- planetary scanner IS2 eScan Open System
- all-in-one PC
- USB external drive (1 TB)
- wireless Zebra barcode reader
- Zebra carbon label printer
- file server (part of IT institutional infrastructure)
Software
- Specify ver 6. (Collection Management System)
- LIMB (digitization software for image capture)
- XnView, image browser and image metadata editor
We also use a number of applications and tools that have been built in house, for file management
Authors
Ana Raquel Cunha, Pedro Arsénio, Rui Figueira
Contributors
This work is built upon the working practices of the RBGE staff.
References
Bandi, Shekar, Mallikarjun Angadi, and J Shivarama. Best Practices in Digitisation: Planning and Workflow Processes, n.d., 8.
Basic Digitization Terminology (D1.b), Digital Collections. Accessed 22 January 2019. http://ulib.hamilton.edu/basic-digitization-terminology-d1b#general_digitization_terminology.
Chapman AD (2005) Principles of Data Quality. Global Biodiversity Information Facility. https://doi.org/10.15468/doc.jrgg-a190.
Creating Archival Master Files, Digital Collections. Accessed 22 January 2019. http://ulib.hamilton.edu/creating-archival-master-files.
Creating Capture Files, Digital Collections. Accessed 22 January 2019. http://ulib.hamilton.edu/creating-capture-files.
Digital Projects Workflow, Digital Collections. Accessed 22 January 2019. http://ulib.hamilton.edu/digital-projects-workflow.
File Naming, Digital Collections. Accessed 22 January 2019. http://ulib.hamilton.edu/file-naming.
Frazier, Cristopher K., John Wall, and Sharon Grant. Initiating a Collection Digitisation Project, Version 1.0. Copenhagen: Global Biodiversity Information Facility, 2008. https://www.gbif.org/document/80574.
Global Plants Initiative (GPI), Kew. Accessed 21 January 2019. https://www.kew.org/science/projects/global-plants-initiative-gpi.
iDigBio. IDigBio Image File Format Requirements and Recommendations, 2014.
iDigBio Wiki. IDigBio Wiki- Specimen Barcode and Labeling Survey Results. Accessed 21 January 2019. https://www.idigbio.org/wiki/index.php/Specimen_Barcode_and_Labeling_Survey_Results.
iDigBio Workflows. Digitization Workflows for Flat Sheets and Packets: IDigBioWorkflows/FlatSheetsDigitizationWorkflows, 2019. https://github.com/iDigBioWorkflows/FlatSheetsDigitizationWorkflows.
Nelson, Gil, Deborah Paul, Greg Riccardi, and Austin Mast. Five Task Clusters That Enable Efficient and Effective Digitization of Biological Collections. ZooKeys 209 (20 July 2012): 19–45. https://doi.org/10.3897/zookeys.209.3135.
Nelson, Gil, Patrick Sweeney, Lisa E. Wallace, Richard K. Rabeler, Dorothy Allard, Herrick Brown, J. Richard Carter, et al. Digitization Workflows for Flat Sheets and Packets of Plants, Algae, and Fungi. Applications in Plant Sciences v.3, nº. 9 (10 September 2015). https://doi.org/10.3732/apps.1500065.
Paul, Deborah, Katja Seltmann, Katja Seltmann, and Katja Seltmann. Biological Collections Data: Best Practices and Trends for Standards, Digitization, and Biodiversity Informatics Literacy for Research Use of Collections Data, 2016.
Revealing the Treasures of Kew’s Herbarium to a Digital World | Kew. Accessed 21 January 2019. https://www.kew.org/blogs/kew-science/revealing-the-treasures-of-kew%E2%80%99s-herbarium-to-a-digital-world. Schmidt, Larry. Digitization of Herbarium Specimens, a Collaborative Project, 2007, 6.
Schmidt, Lawrence. Digitization of Herbarium Specimens, a Collaborative Process, 2007, 7.
Thomas, Hugh. EXIFutils Field Reference Guide, n.d., 178.
Tulig, Melissa, Nicole Tarnowsky, Michael Bevans, Anthony Kirchgessner, and Barbara Thiers. Increasing the Efficiency of Digitization Workflows for Herbarium Specimens. ZooKeys 209 (20 July 2012): 103–13. https://doi.org/10.3897/zookeys.209.3125.
iDigBio. Workflow Modules and Task Lists. Accessed 22 January 2019. https://www.idigbio.org/content/workflow-modules-and-task-lists.
Licence
Citation
Cunha, AR, Arsénio P, Figueira R (2021) LISI Herbarium Digitization Workflow, version 1.0. Instituto Superior de Agronomia, Universidade de Lisboa. 18 pp. https://github.com/PORBIOTA/ISA_DMP/blob/main/LISI_DigitizationWorkflow.pdf
Document Control
Version: 1.0
Changes since last version: N/A
Last Updated: 18 March 2022
Edit This Page
You can suggest changes to this page on our GitHub